4. Advanced example#
In this example, we will show how to run CorrAdjust with several feature types (miRNA-mRNA correlations) and several sample groups (male and female samples). The RNA-seq datasets from lymphoblastoid cell lines (LCLs) for the example are taken from the collection generated by the Geuvadis consortium.
4.1. Loading and normalizing the data#
The below code for reading and normalizing expression data is similar to the previous one with one exception. Specifically, we normalize mRNA and small non-coding RNA (sncRNA) matrices separately, since the datasets come from different experiments and should not be normalized together. After normalization, we concatenate the matrices.
[16]:
# Read the data; both tables have identical index
df_counts_mRNA = pd.read_csv(
"input_data/Geuvadis/raw_counts_mRNA.tsv",
sep="\t", index_col=0
)
df_counts_sncRNA = pd.read_csv(
"input_data/Geuvadis/raw_counts_sncRNA.tsv",
sep="\t", index_col=0
)
# Split into training and test samples
df_counts_mRNA_train = df_counts_mRNA.iloc[::2]
df_counts_mRNA_test = df_counts_mRNA.iloc[1::2]
df_counts_sncRNA_train = df_counts_sncRNA.iloc[::2]
df_counts_sncRNA_test = df_counts_sncRNA.iloc[1::2]
# Normalize mRNA and sncRNA data separately
normalizer = MedianOfRatios()
normalizer.fit(df_counts_mRNA_train)
df_norm_counts_mRNA_train = normalizer.transform(df_counts_mRNA_train)
df_norm_counts_mRNA_test = normalizer.transform(df_counts_mRNA_test)
normalizer = MedianOfRatios()
normalizer.fit(df_counts_sncRNA_train)
df_norm_counts_sncRNA_train = normalizer.transform(df_counts_sncRNA_train)
df_norm_counts_sncRNA_test = normalizer.transform(df_counts_sncRNA_test)
# Concatenate mRNA and sncRNA data
df_norm_counts_train = pd.concat(
[df_norm_counts_sncRNA_train, df_norm_counts_mRNA_train],
axis=1
)
df_norm_counts_test = pd.concat(
[df_norm_counts_sncRNA_test, df_norm_counts_mRNA_test],
axis=1
)
# Log-transform
df_data_train = np.log2(df_norm_counts_train + 1)
df_data_test = np.log2(df_norm_counts_test + 1)
display(df_data_train)
| tRF-33-79MP9P9NH57SD3 | iso-23-2HOMKBFPDP | rRF-18-WLLINPD4 | tRF-33-Z3M8ZLSSXUOLD2 | iso-22-X2EUIRIKQ | iso-22-BQ8DQWM4H | iso-24-YEQOW4HK2E | yRF-27-KR2Y4BZUNJQ | tRF-18-HR6HFRD2 | iso-24-VY2ZSR672F | ... | ENSG00000198712.1 | ENSG00000228253.1 | ENSG00000198899.2 | ENSG00000198938.2 | ENSG00000198840.2 | ENSG00000212907.2 | ENSG00000198886.2 | ENSG00000198786.2 | ENSG00000198695.2 | ENSG00000198727.2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TSI_NA20508.1.MI | 15.375467 | 15.768876 | 14.386797 | 11.509986 | 17.077093 | 16.831354 | 16.979418 | 15.446145 | 17.407458 | 15.867370 | ... | 16.957235 | 12.100007 | 16.197138 | 17.012233 | 13.547551 | 13.994002 | 17.536592 | 16.437674 | 14.407458 | 15.823431 |
| FIN_HG00315.2.MI | 17.004050 | 14.824433 | 15.759253 | 13.413163 | 14.623674 | 14.236777 | 15.006757 | 14.406483 | 15.180716 | 14.415009 | ... | 16.971874 | 12.258090 | 15.962215 | 17.203759 | 13.432169 | 13.696331 | 17.444533 | 16.435032 | 15.052151 | 16.405424 |
| FIN_HG00377.2.MI | 17.363959 | 16.194561 | 15.104459 | 11.785276 | 15.627775 | 15.123750 | 15.992820 | 15.160283 | 14.282834 | 15.429691 | ... | 17.195135 | 12.745818 | 16.419661 | 17.434837 | 13.754321 | 13.820417 | 17.623701 | 16.587501 | 15.150230 | 16.501734 |
| CEU_NA07056.1.MI | 17.097118 | 17.129173 | 17.445657 | 14.503363 | 16.753292 | 14.451858 | 15.225481 | 14.832857 | 15.499062 | 15.173789 | ... | 18.067899 | 12.790246 | 17.218922 | 18.242186 | 14.478591 | 14.349128 | 18.692826 | 17.028353 | 15.858036 | 16.719261 |
| YRI_NA19119.3.MI | 16.795022 | 17.570451 | 15.459582 | 11.381865 | 17.177996 | 15.552042 | 16.201064 | 15.789840 | 17.553967 | 16.495589 | ... | 16.224141 | 12.177533 | 15.455781 | 16.560902 | 13.195824 | 13.352683 | 16.708912 | 15.811658 | 14.117363 | 15.174058 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| GBR_HG01791.2.MI | 16.126779 | 15.715584 | 14.766975 | 15.139796 | 16.288074 | 16.552333 | 15.605472 | 14.096111 | 16.977521 | 15.361669 | ... | 16.720027 | 12.270003 | 15.723737 | 16.764922 | 13.368546 | 13.347992 | 17.085581 | 16.192440 | 14.622754 | 15.805635 |
| FIN_HG00362.3.MI | 17.121267 | 16.418557 | 13.368475 | 14.779898 | 16.243674 | 15.027084 | 15.819105 | 15.377365 | 13.327610 | 15.522643 | ... | 16.910549 | 12.079542 | 15.968046 | 17.013613 | 13.524998 | 13.623903 | 17.297789 | 16.082245 | 14.738878 | 15.962373 |
| CEU_NA07000.1.MI | 16.410787 | 17.711002 | 17.774399 | 12.880337 | 17.419655 | 16.219216 | 16.623148 | 15.430145 | 17.526397 | 16.129083 | ... | 17.297790 | 11.366524 | 16.511681 | 17.707241 | 14.237225 | 14.045978 | 18.140950 | 16.570033 | 15.179156 | 16.700573 |
| YRI_NA18861.4.MI | 15.755783 | 16.457623 | 15.142608 | 11.822476 | 16.368148 | 16.651518 | 16.597095 | 15.635380 | 16.886426 | 16.344094 | ... | 13.245564 | 9.010921 | 12.461696 | 13.331866 | 10.670205 | 10.269527 | 14.117214 | 14.357536 | 12.045723 | 13.455430 |
| GBR_HG00237.4.MI | 13.614071 | 14.934466 | 11.768121 | 13.726310 | 14.688703 | 16.593015 | 16.395290 | 15.619196 | 14.995440 | 15.242237 | ... | 17.009939 | 12.551017 | 15.668342 | 17.099195 | 13.859134 | 13.359202 | 16.929224 | 15.904793 | 14.359246 | 15.802043 |
202 rows × 15770 columns
Fene annotation file also follows the same format but now we have two feature types (sncRNA and mRNA):
[17]:
df_feature_ann = pd.read_csv(
"input_data/Geuvadis/gene_annotation.tsv",
sep="\t", index_col=0
)
display(df_feature_ann)
| feature_name | feature_type | |
|---|---|---|
| feature_id | ||
| tRF-33-79MP9P9NH57SD3 | tRF-33-79MP9P9NH57SD3 | sncRNA |
| iso-23-2HOMKBFPDP | iso-23-2HOMKBFPDP&hsa-miR-191-5p|0|0 | sncRNA |
| rRF-18-WLLINPD4 | rRF-18-WLLINPD4 | sncRNA |
| tRF-33-Z3M8ZLSSXUOLD2 | tRF-33-Z3M8ZLSSXUOLD2 | sncRNA |
| iso-22-X2EUIRIKQ | iso-22-X2EUIRIKQ&hsa-miR-146a-5p|0|0 | sncRNA |
| ... | ... | ... |
| ENSG00000212907.2 | MT-ND4L | mRNA |
| ENSG00000198886.2 | MT-ND4 | mRNA |
| ENSG00000198786.2 | MT-ND5 | mRNA |
| ENSG00000198695.2 | MT-ND6 | mRNA |
| ENSG00000198727.2 | MT-CYB | mRNA |
15770 rows × 2 columns
Finally, we read a table with annotation of samples (male and female). Note that the group column could have an arbitrary number of different values.
[18]:
df_samp_ann = pd.read_csv(
"input_data/Geuvadis/sample_annotation.tsv",
sep="\t", index_col=0
)
df_samp_ann_train = df_samp_ann.iloc[::2]
df_samp_ann_test = df_samp_ann.iloc[1::2]
display(df_samp_ann_train)
| group | |
|---|---|
| sample | |
| TSI_NA20508.1.MI | female |
| FIN_HG00315.2.MI | female |
| FIN_HG00377.2.MI | female |
| CEU_NA07056.1.MI | female |
| YRI_NA19119.3.MI | male |
| ... | ... |
| GBR_HG01791.2.MI | male |
| FIN_HG00362.3.MI | female |
| CEU_NA07000.1.MI | female |
| YRI_NA18861.4.MI | female |
| GBR_HG00237.4.MI | female |
202 rows × 1 columns
As a reference feature pair sets, we will use experimentally validated targets from TarBase v9. Note that targets are available only for canonical isomiRs (isomiRs with |0|0 suffix), while expression data has both canonical and non-canonical ones, as well as other non-isomiR sncRNAs.
[19]:
ref_feature_colls = {
"TarBase": {
"path": "input_data/GMT_files/TarBase.gmt",
# We expect targets among negative miRNA-mRNA correlations
"sign": "negative",
"feature_pair_types": ["sncRNA-mRNA"],
# Good default value for miRNA-mRNA analysis
"high_corr_frac": 0.05
}
}
4.2. Training the model#
Now we create and fit the model, passing sample annotation as an additional argument to fit.
[20]:
model = CorrAdjust(
df_feature_ann,
ref_feature_colls,
"out_data/Geuvadis"
)
model.fit(
df_data_train,
df_samp_ann=df_samp_ann_train,
# Set n_PCs value from paper
n_PCs=17
)
2025-02-28 20:01:28.968561 | Loading TarBase reference collection...
2025-02-28 20:01:31.982782 | Computing PCA...
2025-02-28 20:01:33.001837 | Starting PC optimization...
100%|██████████████████████████████████████████████████████████████| 154/154 [24:48<00:00, 9.67s/it]
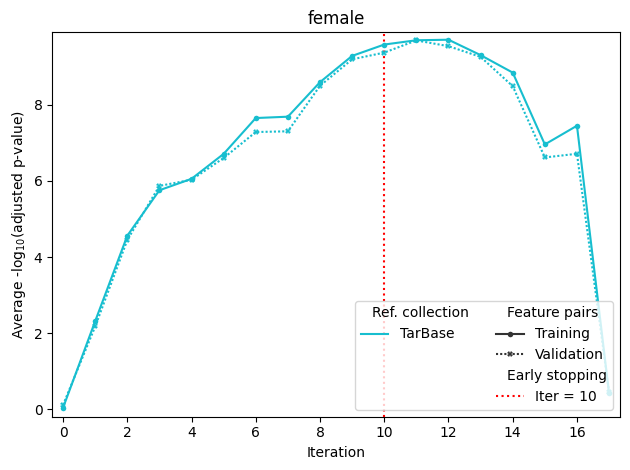
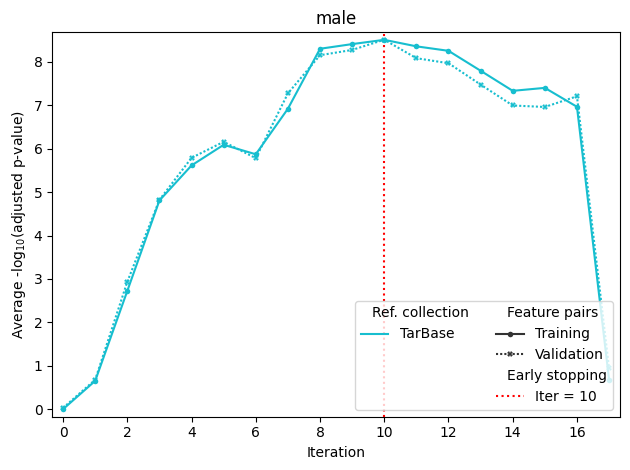
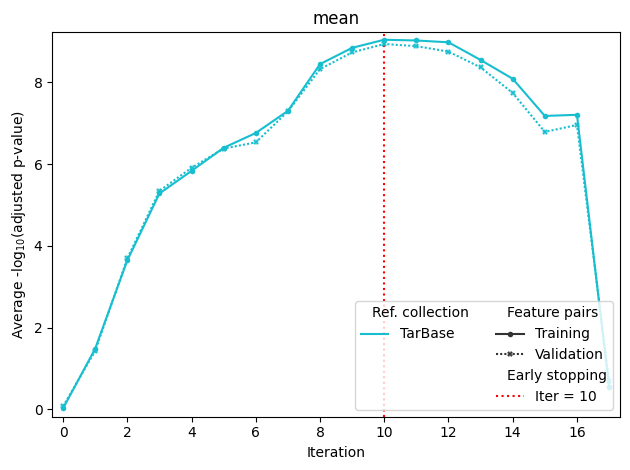
If sample annotation with more than one group is passed to fit, it will produce separate optimization plots for each group. The last plot stands for mean scores (in this case, mean of male and female scores). Mean training score is used for maximization, and mean validation score is used for early stopping.
4.3. Using the trained model#
Calls of all downstream methods should include sample annotation and sample group name. To avoid code duplication, this can be done in a loop.
[21]:
for samp_group in ["female", "male"]:
feature_scores = model.compute_feature_scores(
df_data_test,
df_samp_ann=df_samp_ann_test,
samp_group=samp_group
)
# This attribute adds title in top-left corners of plots
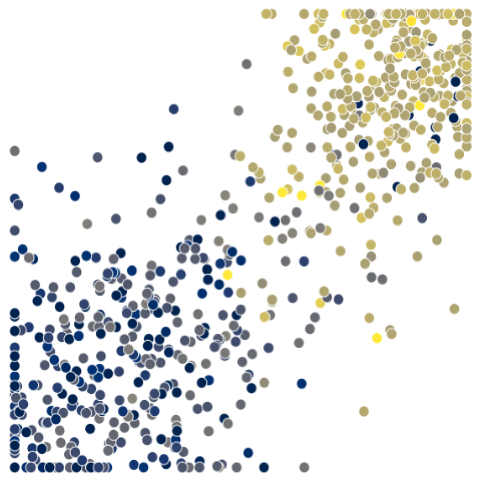
model.title = f"Sex = {samp_group}"
model.make_volcano_plot(
feature_scores,
f"volcano.test_samples.{samp_group}.png",
annotate_features=10,
# Use formatter function to shorten isomiR names
feature_name_fmt=lambda name: name.split("hsa-")[1].split("|")[0]
)
model.make_corr_distr_plot(
df_data_test,
f"corr_distr.test_samples.{samp_group}.png",
df_samp_ann=df_samp_ann_test,
samp_group=samp_group
)
model.export_corrs(
df_data_test,
f"corrs.test_samples.{samp_group}.tsv",
df_samp_ann=df_samp_ann_test,
samp_group=samp_group
)
# These lines just show a representative chunk of the file
corrs = pd.read_csv(
f"out_data/Geuvadis/corrs.test_samples.{samp_group}.tsv",
sep="\t", nrows=100000
)
print(f"Chunk of corrs.test_samples.{samp_group}.tsv:")
display(corrs.loc[corrs["TarBase_flag"] != -1].head(10))
2025-02-28 20:26:24.436736 | Computing raw correlations for female...
2025-02-28 20:26:26.703805 | Computing corrected correlations for female...
2025-02-28 20:26:36.252894 | Computing raw correlations for female...
2025-02-28 20:26:38.543961 | Computing corrected correlations for female...
2025-02-28 20:26:48.916197 | Computing raw correlations for female...
2025-02-28 20:26:51.184528 | Computing corrected correlations for female...
2025-02-28 20:26:56.129525 | Starting export to file...
100%|██████████████████████████████████████████████████████████████| 125/125 [05:22<00:00, 2.58s/it]
Chunk of corrs.test_samples.female.tsv:
| feature_id1 | feature_id2 | feature_name1 | feature_name2 | feature_type1 | feature_type2 | corr_clean | pvalue_clean | corr_raw | pvalue_raw | TarBase_flag | TarBase_trainval | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1587 | iso-22-VFPFU47YP | ENSG00000082701.17 | iso-22-VFPFU47YP&hsa-miR-15a-5p|0|0 | GSK3B | sncRNA | mRNA | -0.715228 | 3.162188e-31 | 0.134124 | 0.057662 | 1 | 0 |
| 4837 | iso-23-U0XZH3PK0H | ENSG00000082701.17 | iso-23-U0XZH3PK0H&hsa-miR-20a-5p|0|0 | GSK3B | sncRNA | mRNA | -0.683877 | 1.117633e-27 | 0.244307 | 0.000474 | 1 | 0 |
| 6494 | iso-23-20XYYWPK0H | ENSG00000131023.13 | iso-23-20XYYWPK0H&hsa-miR-93-5p|0|0 | LATS1 | sncRNA | mRNA | -0.675366 | 8.629332e-27 | 0.179440 | 0.010808 | 1 | 0 |
| 6997 | iso-22-83PI02EZN | ENSG00000118513.19 | iso-22-83PI02EZN&hsa-miR-148a-3p|0|0 | MYB | sncRNA | mRNA | -0.673046 | 1.488833e-26 | -0.276047 | 0.000073 | 1 | 1 |
| 7477 | iso-23-9P9Z35ZQDX | ENSG00000082701.17 | iso-23-9P9Z35ZQDX&hsa-miR-142-3p|0|0 | GSK3B | sncRNA | mRNA | -0.670901 | 2.454460e-26 | 0.186854 | 0.007907 | 0 | 1 |
| 7712 | iso-23-J42LSNSO9 | ENSG00000109171.15 | iso-23-J42LSNSO9&hsa-miR-301a-3p|0|0 | SLAIN2 | sncRNA | mRNA | -0.669953 | 3.056737e-26 | 0.061544 | 0.385438 | 1 | 0 |
| 9829 | iso-22-URIVHM63P | ENSG00000060237.18 | iso-22-URIVHM63P&hsa-miR-660-5p|0|0 | WNK1 | sncRNA | mRNA | -0.662119 | 1.820237e-25 | 0.173901 | 0.013552 | 1 | 1 |
| 10006 | iso-23-U0XZH3PK0H | ENSG00000109171.15 | iso-23-U0XZH3PK0H&hsa-miR-20a-5p|0|0 | SLAIN2 | sncRNA | mRNA | -0.661582 | 2.052673e-25 | 0.167919 | 0.017184 | 1 | 1 |
| 10875 | iso-23-J42LSNSO9 | ENSG00000082701.17 | iso-23-J42LSNSO9&hsa-miR-301a-3p|0|0 | GSK3B | sncRNA | mRNA | -0.658831 | 3.787756e-25 | 0.290319 | 0.000029 | 1 | 1 |
| 11858 | iso-23-J424HNSO9 | ENSG00000048405.10 | iso-23-J424HNSO9&hsa-miR-301b-3p|0|0 | ZNF800 | sncRNA | mRNA | -0.655913 | 7.202982e-25 | 0.169388 | 0.016221 | 1 | 0 |
2025-02-28 20:32:19.835571 | Computing raw correlations for male...
2025-02-28 20:32:22.327870 | Computing corrected correlations for male...
2025-02-28 20:32:32.340275 | Computing raw correlations for male...
2025-02-28 20:32:34.848362 | Computing corrected correlations for male...
2025-02-28 20:32:45.141563 | Computing raw correlations for male...
2025-02-28 20:32:47.713209 | Computing corrected correlations for male...
2025-02-28 20:32:52.741026 | Starting export to file...
100%|███████████████████████████████████████████████████████████████| 125/125 [05:14<00:00, 2.52s/it]
Chunk of corrs.test_samples.male.tsv:
| feature_id1 | feature_id2 | feature_name1 | feature_name2 | feature_type1 | feature_type2 | corr_clean | pvalue_clean | corr_raw | pvalue_raw | TarBase_flag | TarBase_trainval | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 21390 | iso-23-20XYYWPK0H | ENSG00000130338.13 | iso-23-20XYYWPK0H&hsa-miR-93-5p|0|0 | TULP4 | sncRNA | mRNA | -0.655115 | 8.576050e-25 | -0.066628 | 0.347335 | 1 | 1 |
| 21892 | iso-21-42NBQRZ00 | ENSG00000009307.16 | iso-21-42NBQRZ00&hsa-miR-151a-3p|0|0 | CSDE1 | sncRNA | mRNA | -0.654322 | 1.019516e-24 | -0.175774 | 0.012562 | 0 | 1 |
| 29488 | iso-23-U0XZH3PK0H | ENSG00000082701.17 | iso-23-U0XZH3PK0H&hsa-miR-20a-5p|0|0 | GSK3B | sncRNA | mRNA | -0.643953 | 9.335504e-24 | 0.120305 | 0.088911 | 1 | 0 |
| 32337 | iso-21-42NBQRZ00 | ENSG00000138593.9 | iso-21-42NBQRZ00&hsa-miR-151a-3p|0|0 | SECISBP2L | sncRNA | mRNA | -0.640631 | 1.863912e-23 | -0.189063 | 0.007188 | 0 | 1 |
| 34594 | iso-22-J424IB1JI | ENSG00000118058.23 | iso-22-J424IB1JI&hsa-miR-130b-3p|0|0 | KMT2A | sncRNA | mRNA | -0.638107 | 3.134571e-23 | 0.211219 | 0.002613 | 1 | 1 |
| 41222 | iso-23-20XZ03PK0H | ENSG00000102908.22 | iso-23-20XZ03PK0H&hsa-miR-17-5p|0|0 | NFAT5 | sncRNA | mRNA | -0.631484 | 1.198429e-22 | 0.170516 | 0.015514 | 1 | 1 |
| 42711 | iso-21-W05I2PWPE | ENSG00000009307.16 | iso-21-W05I2PWPE&hsa-miR-151a-5p|0|0 | CSDE1 | sncRNA | mRNA | -0.630074 | 1.587678e-22 | -0.113037 | 0.110109 | 1 | 1 |
| 44583 | iso-23-20XZ03PK0H | ENSG00000110367.13 | iso-23-20XZ03PK0H&hsa-miR-17-5p|0|0 | DDX6 | sncRNA | mRNA | -0.628444 | 2.194175e-22 | 0.084632 | 0.232272 | 1 | 0 |
| 46898 | iso-22-XKVL7YXYQ | ENSG00000118058.23 | iso-22-XKVL7YXYQ&hsa-let-7i-5p|0|0 | KMT2A | sncRNA | mRNA | -0.626423 | 3.267926e-22 | 0.112801 | 0.110857 | 1 | 1 |
| 49526 | iso-23-U0XZH3PK0H | ENSG00000110367.13 | iso-23-U0XZH3PK0H&hsa-miR-20a-5p|0|0 | DDX6 | sncRNA | mRNA | -0.624238 | 5.011761e-22 | 0.033899 | 0.632831 | 1 | 1 |
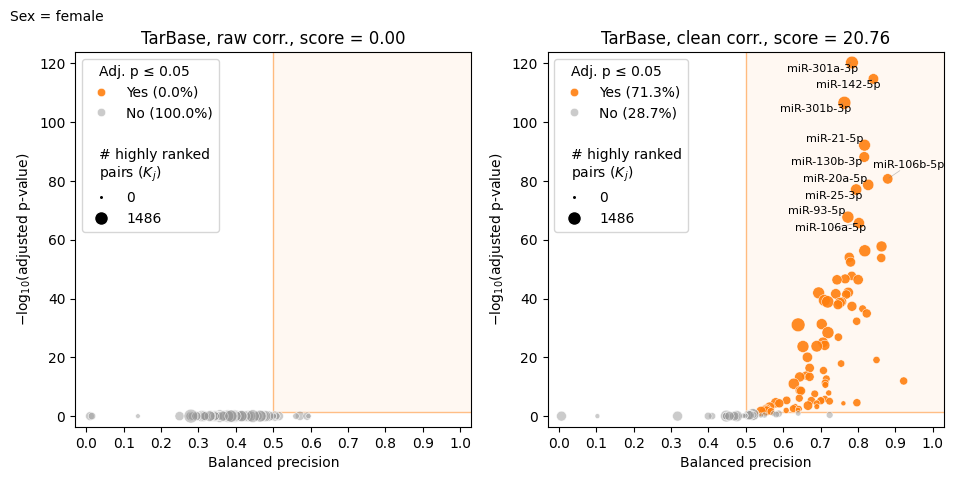
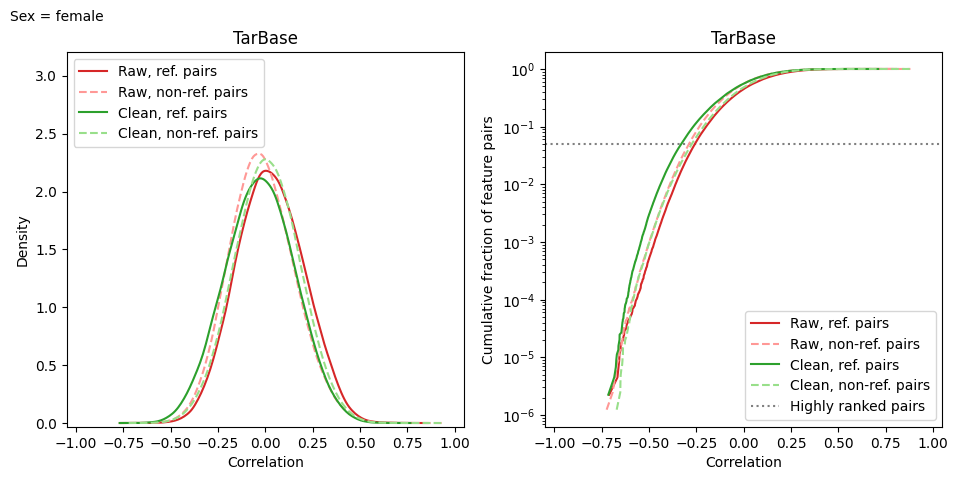
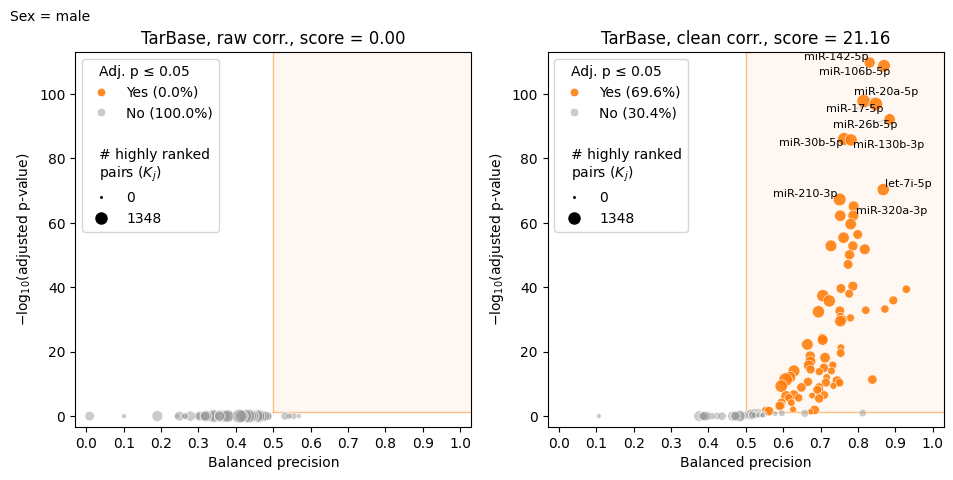
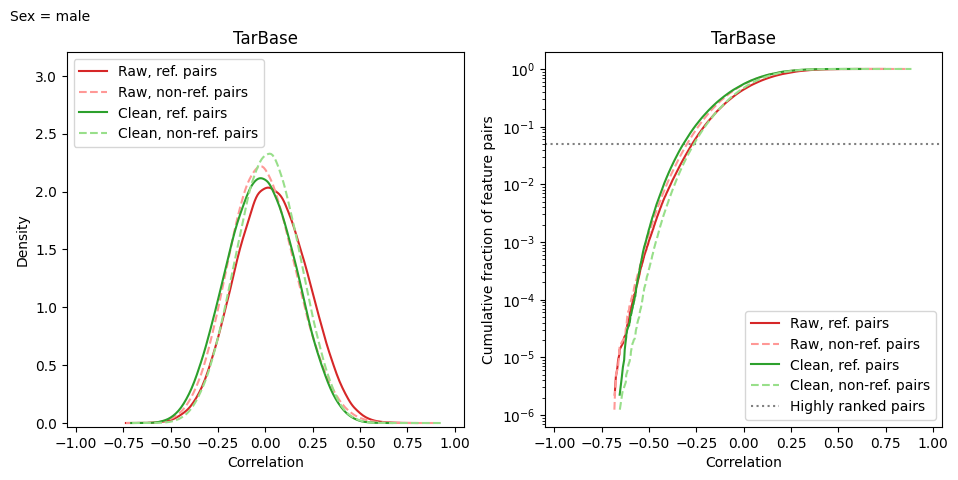
The only method that doesn’t require group name is transform, since all samples are cleaned together.
[22]:
df_data_test_clean, df_rsquareds = model.transform(
df_data_test,
df_samp_ann_test
)
display(df_data_test_clean)
| tRF-33-79MP9P9NH57SD3 | iso-23-2HOMKBFPDP | rRF-18-WLLINPD4 | tRF-33-Z3M8ZLSSXUOLD2 | iso-22-X2EUIRIKQ | iso-22-BQ8DQWM4H | iso-24-YEQOW4HK2E | yRF-27-KR2Y4BZUNJQ | tRF-18-HR6HFRD2 | iso-24-VY2ZSR672F | ... | ENSG00000198712.1 | ENSG00000228253.1 | ENSG00000198899.2 | ENSG00000198938.2 | ENSG00000198840.2 | ENSG00000212907.2 | ENSG00000198886.2 | ENSG00000198786.2 | ENSG00000198695.2 | ENSG00000198727.2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CEU_NA12812.1.MI | -0.620065 | -0.310726 | -0.249793 | 1.290901 | 0.281442 | -0.050428 | 0.527796 | -0.401979 | -0.772192 | -0.512847 | ... | 0.158439 | 0.173913 | 0.427129 | 0.534144 | 0.454318 | 0.439413 | 0.549146 | 0.316825 | 0.381061 | 0.078790 |
| CEU_NA12749.1.MI | -1.106119 | -0.628111 | -1.249008 | 0.616152 | 0.206401 | -0.108065 | 0.103019 | 0.038530 | 1.169227 | -0.064189 | ... | -0.344318 | -0.269753 | -0.155804 | -0.453415 | -0.155392 | -0.505204 | -0.384939 | -0.579743 | -0.510531 | -0.332724 |
| TSI_NA20510.3.MI | 0.195529 | -0.151787 | 0.250982 | -0.509563 | 0.062368 | -0.322359 | -0.387207 | 0.716692 | 0.794845 | -0.685646 | ... | 0.132211 | 0.255880 | 0.054310 | -0.062379 | 0.110532 | 0.161819 | 0.200947 | 0.060922 | 0.095826 | -0.142283 |
| GBR_HG00155.1.MI | -0.053329 | -0.300658 | 0.170739 | -0.383377 | -0.068466 | -0.254343 | -0.439967 | -0.292031 | 1.943921 | -0.449839 | ... | -0.092572 | -0.054234 | 0.190476 | 0.035316 | 0.234884 | 0.145346 | 0.030836 | -0.008509 | -0.025680 | 0.144530 |
| FIN_HG00350.3.MI | 0.525551 | -0.086287 | 0.157886 | -0.400232 | 0.006228 | -0.445866 | -0.210717 | 0.212302 | 1.625556 | -0.428203 | ... | -0.038814 | -0.153922 | -0.146719 | 0.017803 | 0.096110 | -0.301353 | 0.002004 | -0.402903 | -0.169281 | -0.109961 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| CEU_NA12778.1.MI | 0.581613 | -0.307854 | 1.827755 | 0.375388 | 0.559242 | 0.283521 | -0.132107 | 0.957126 | 0.674212 | -0.213702 | ... | 0.122245 | -0.301193 | 0.274027 | 0.291671 | -0.014586 | -0.008767 | 0.404175 | 0.418945 | 0.170228 | 0.276137 |
| TSI_NA20761.1.MI | 0.427250 | -0.000609 | 1.854137 | -0.967633 | 0.286078 | -0.553838 | -0.271268 | 0.738555 | 0.066126 | -0.495017 | ... | -0.095754 | -1.027643 | -0.271515 | -0.459521 | -0.555034 | -0.464335 | 0.236649 | 0.014550 | 0.192458 | -0.273718 |
| FIN_HG00308.3.MI | 0.049832 | 0.064105 | 0.216882 | 0.504403 | -0.077997 | -0.153369 | -0.179680 | -0.236491 | 0.524470 | 0.310773 | ... | 0.224791 | 0.431358 | 0.144727 | 0.234761 | 0.124680 | 0.232421 | 0.155974 | -0.221077 | -0.224894 | -0.307741 |
| CEU_NA12399.7.MI | 0.996266 | -0.470806 | -0.691879 | 0.213296 | -0.218509 | 0.302927 | 0.066839 | 0.112897 | -1.323507 | 0.208681 | ... | -0.306433 | 0.255695 | -0.451077 | -0.298684 | -0.363276 | -0.000983 | -0.401184 | -0.278655 | -0.341041 | -0.359523 |
| YRI_NA19144.4.MI | -0.631015 | -0.555298 | -2.123311 | -0.045040 | -0.326791 | -0.092634 | -0.241750 | 0.372753 | 0.076177 | -0.711939 | ... | -0.074233 | -1.253004 | -0.406206 | 0.013982 | 0.171947 | -0.449277 | -0.364824 | -0.354861 | 0.004093 | -0.504313 |
201 rows × 15770 columns